Illumina has introduced the possibility of 300bp sequencing for 16sRNA; this has encouraged a lot of researchers to sequence both V3 and V4 regions and have a more accurate microbiome profiles. The alternative methods that was excercised so far and Patric Scloss advocate is the using the 250bp reads for sequencing the only V3 region.
However, many people have complained about the new illumina protocol for sequencing V3 and V4. The main argument against sequencing two regions with longer reads is the quality of 300 bp reads. Pat Schloss has shown that the quality of base pairs after ~250 is same and the extera 50bp (at each end) is junk. So it is not going to cover entirly both V3 and V4; and the low quality base pair increase the number of unique sequences which will results in higher number of spicies. His methods looks more reasonable because the V3 reagion is shorter than 500bp and reads that lay down on the region V3 have an overal higher quality. Mothur never finishes the analysis because the corrlation matrix is extremely huge.
Here is a pipleline to get advantage of the most.
I have just put it in gist gist
|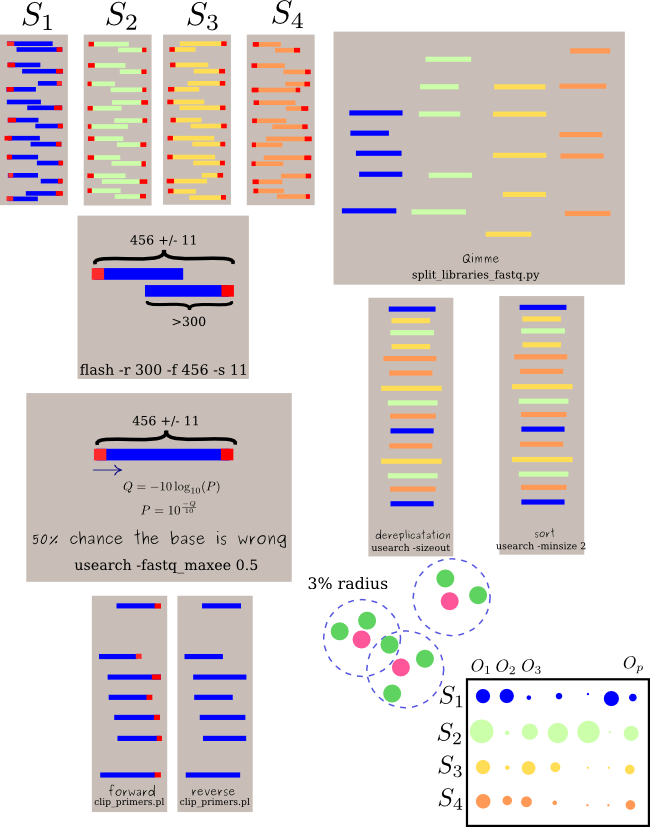 |
|
Note:
- I am going to update the page - please help me with your valuable comments.
- hopefully I will find time to keep it clean and readable.
FLASH vs make.contig
The quality of mothur assembly (make.contig) looks lower than FLASH. By quality I mean the reasonable lenght for the contig lenght is much higher than forward_lenght+reverse_length; I know mothur has a lot of smart filtering ways to exclude these bad contiges; but based on my analysis seems the contigs made by FLASH are more at the first place.
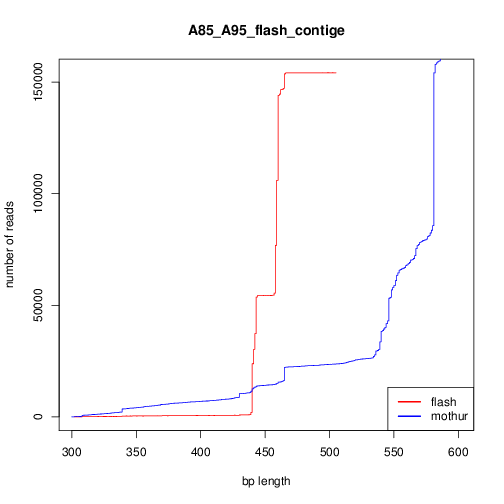
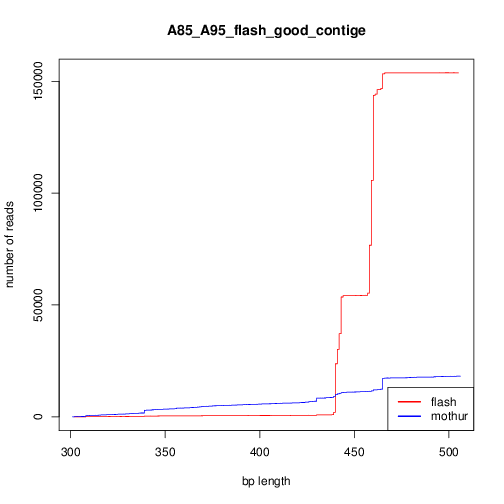
Left side figure, shows number of contigs versus their lenght just after the assembly. The right sides, is the same, but by excluding reads longer than 506 (460+46).
Comments
comments powered by Disqus